
Magdalena Maslon - RNA biology
Science: Cellular functions rely on the precise coordination of gene expression, while the disruption of this process forms the basis of many developmental diseases. Our group studies basic gene expression mechanisms in mammalian development.
Ethos: We value excellence and prioritize transparency and kindness
Tools: stem cell models, mouse models, high-throughput sequencing technologies, CRISPR-Cas9-mediated genome editing, siRNA screens, bioinformatics
Scientific summary:
Our genomes are highly regulated, and their expression can respond to a variety of environmental cues. For example, during development, gene expression programs drive the generation of thousands of cell types in our bodies from a small population of pluripotent cells.
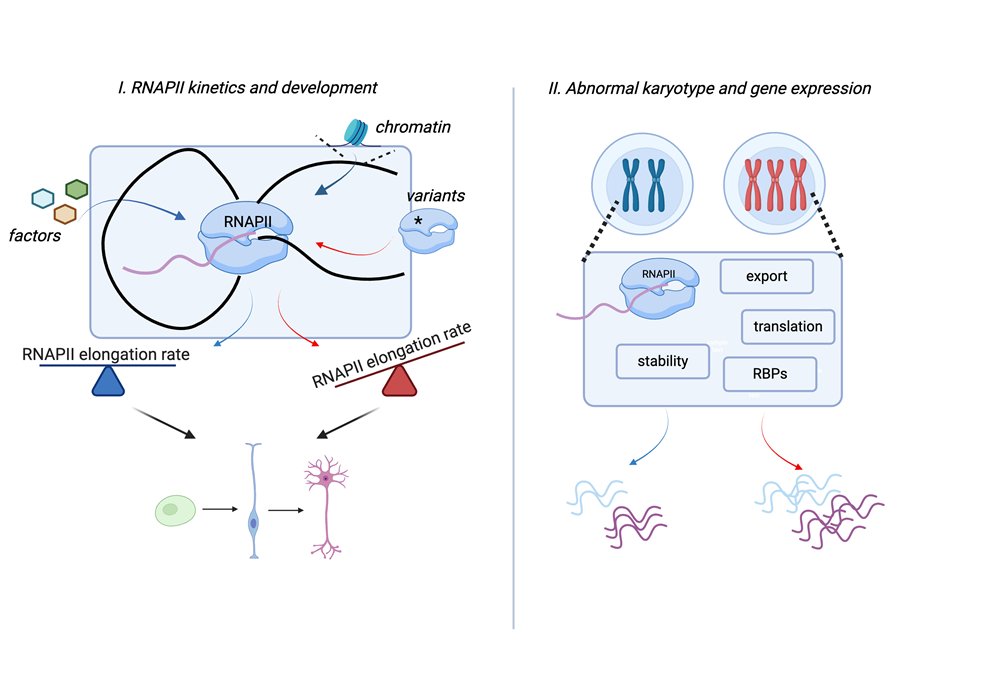
The key enzyme that decodes eukaryotic genomes and that gives rise to all mRNAs and many non-coding RNAs is RNA polymerase II (RNAPII). RNAPII elongation rates are highly dynamic and vary between genes and across the gene body. Due to the co-transcriptional nature of RNA processing, RNAPII rates regulate processes such as alternative splicing and polyadenylation. Thus, changes in RNAPII speed can modify the composition of the transcriptome.
Previously, I generated an in vivo mouse model of RNAPII (Polr2aR749H) with a lower elongation rate (“slow” RNAPII). I found that “slow” RNAPII results in embryonic lethality in mice, impairs the differentiation of embryonic stem cells to the neural lineage and leads to the downregulation of long transcripts in neurons. This work revealed that RNAPII kinetics serve as important regulatory step in development, but we are far from understanding the mechanism of transcription elongation regulation during differentiation and how its mis-regulation contributes to disease.
1. Discover the key mechanisms that underlie the regulation of nascent RNAPII transcription. Specifically, we aim to understand RNAPII elongation rate regulation in development and whether its inappropriate regulation can lead to disease.
2. Understand how fundamental gene expression programs are altered in cells with abnormal genomes. We focus on Trisomy 21 (T21) and how T21 changes RNAPII transcription in fundamental ways to increase RNA output.
1 joint first author
Haward F1, Maslon MM1, Yeyati PL1, Bellora N, Hansen JN, Aitken S, Lawson J, von Kriegsheim A, Wachten D, Mill P, Adams IR, Caceres JF. Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. Elife. 2021 Aug 2;10:e65104. doi: 10.7554/eLife.65104. PMID: 34338635
Maslon MM, Braunschweig U, Aitken S, Mann AR, Kilanowski F, Hunter CJ, Blencowe BJ, Kornblihtt AR, Adams IR, Cáceres JF. A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J. 2019 May 2;38(9):e101244. doi: 10.15252/embj.2018101244.
Maslon MM1, Heras SR1, Bellora N, Eyras E, Cáceres JF. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife. 2014 May 6;3:e02028. doi: 10.7554/eLife.02028.

dr Magdalena Maslon


