Research on the gut microbiome of COVID-19 patients, published in Frontiers in Microbiology by a team from the Małopolska Centre of Biotechnology, reveals how the microbiome influences the course of the disease in hospitalized patients
Effects of COVID-19 on the gastrointestinal tract
The COVID-19 pandemic caused by SARS-CoV-2 has caused a wide range of clinical manifestations, with respiratory symptoms being the most common. However, emerging evidence suggests that the gastrointestinal tract is also affected by the virus. Angiotensin-converting enzyme 2 (ACE2), a key receptor for the SARS-CoV-2 virus, is abundant in the ileum and colon. The SARS-CoV-2 virus has been detected in gastrointestinal tract tissues and stool samples, even in cases of negative SARS-Cov-2 tests in the respiratory tract. Gastrointestinal tract-related symptoms were associated with an increased risk of intensive care unit hospitalization and mortality.
The role of the gut microbiome
The gut microbiome, a complex ecosystem of about 40 billion bacteria, plays a key role in immune and metabolic pathways. Dysbiosis of the gut microbiota, characterized by loss of beneficial microorganisms and reduced microbial diversity, has been observed in patients with COVID-19, potentially contributing to the severity of the disease.
A study of the gut microbiome in COVID-19
In a publication published in Frontiers in Microbiology, researchers analyzed the gut microbiome of 204 patients hospitalized for severe COVID-19. The aim of the study was to track changes in microbial composition during hospitalization and relate these changes to clinical procedures (antibiotic administration, ICU admission, hospitalization outcome: survival or death). The predictive potential of the gut microbiome for the prognosis of COVID-19 was evaluated, and the differential effects of clinical parameters, patient baseline data (gender, age, BMI) and microbiome on the accuracy of predicting the outcome of hospitalization (ICU referral, survival/death) were demonstrated. It was shown that microbiome data have higher reliability in predicting patient outcomes compared to clinical or baseline data.
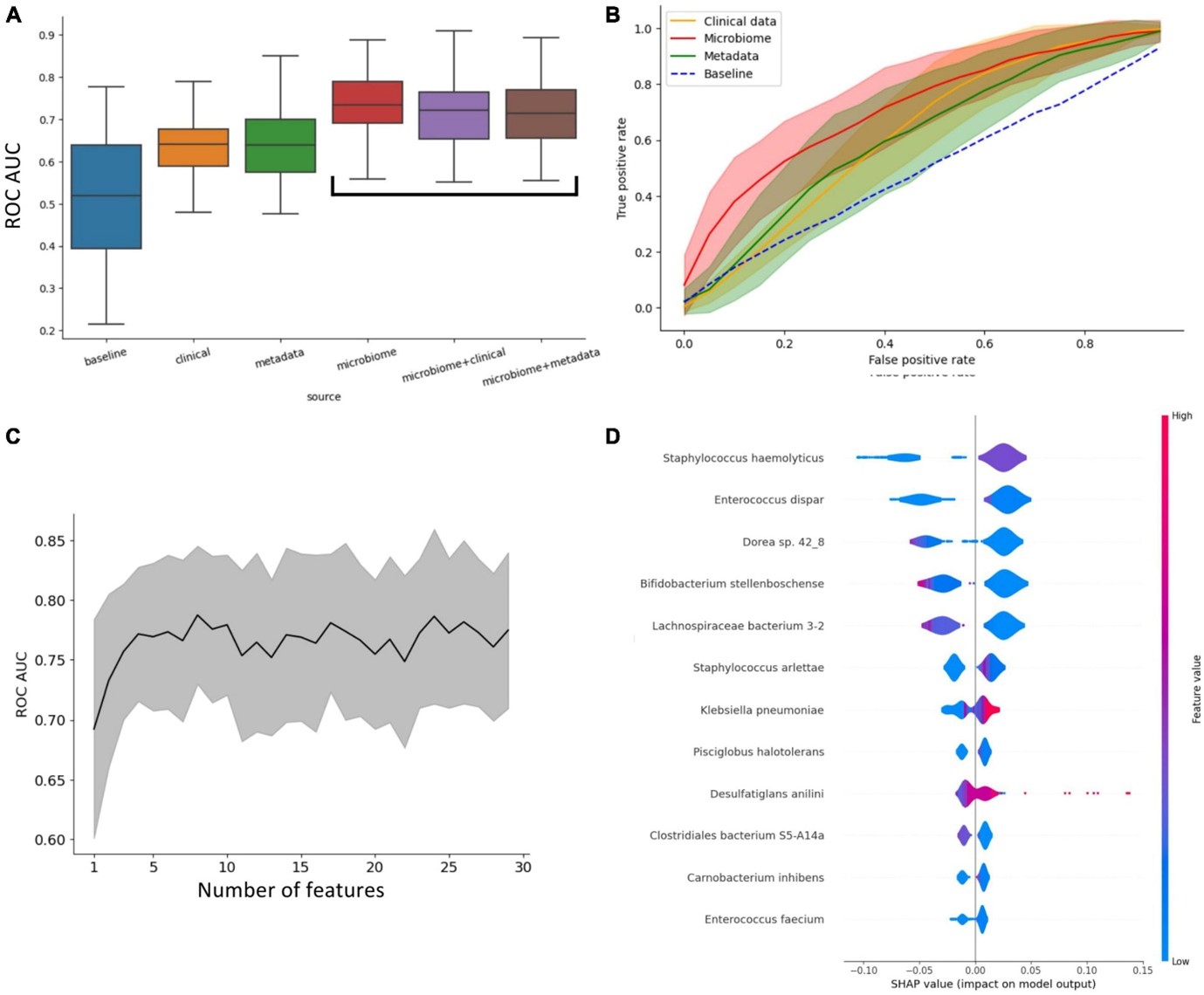
Figure 1. Effect of clinical parameters, patient baseline data (gender, age, BMI) and microbiome on the accuracy of predicting hospitalization outcome (ICU vs. non-ICU referral). (A) Impact of different types of data on the ability to predict hospitalization outcome. Microbiome data significantly improve the performance of classifiers. (B) ROC curve of classifiers grouped by data type. (C) Increased microbiome diversity does not improve ROC-AUC results except for 7 relevant species. (D) Shapley values for significant species.
Comparison of microbiome sequencing methods
Recent studies on the gut microbiome of COVID-19 patients have used two main sequencing methods: 16S rRNA gene sequencing and deep shotgun metagenomic sequencing. The former method is more economical and suitable for large sample sets, but does not provide as precise information on bacterial genera as deep shotgun sequencing. The latter is more accurate but expensive, limiting its widespread use in clinical trials. It turns out that shallow shotgun sequencing, which is less expensive than deep sequencing, provides equally accurate results on key aspects of the microbiome, making it a promising tool for use in clinical practice, especially in the context of COVID-19 management.
Shallow shotgun sequencing as an alternative
Dr. Tomasz Kościółek's team demonstrated that shallow shotgun sequencing is a viable and cost-effective diagnostic alternative to deep sequencing in clinical settings. They observed high coverage of species identified by shallow and deep sequencing. Deeper sequencing revealed significantly more species, but all but five species identified by shallow sequencing were also detected by deep sequencing. In addition, we showed that although species abundance was not perfectly matched between shallow and deep sequencing, the species abundance hierarchy, even at low abundances of less than 1%, was well preserved.
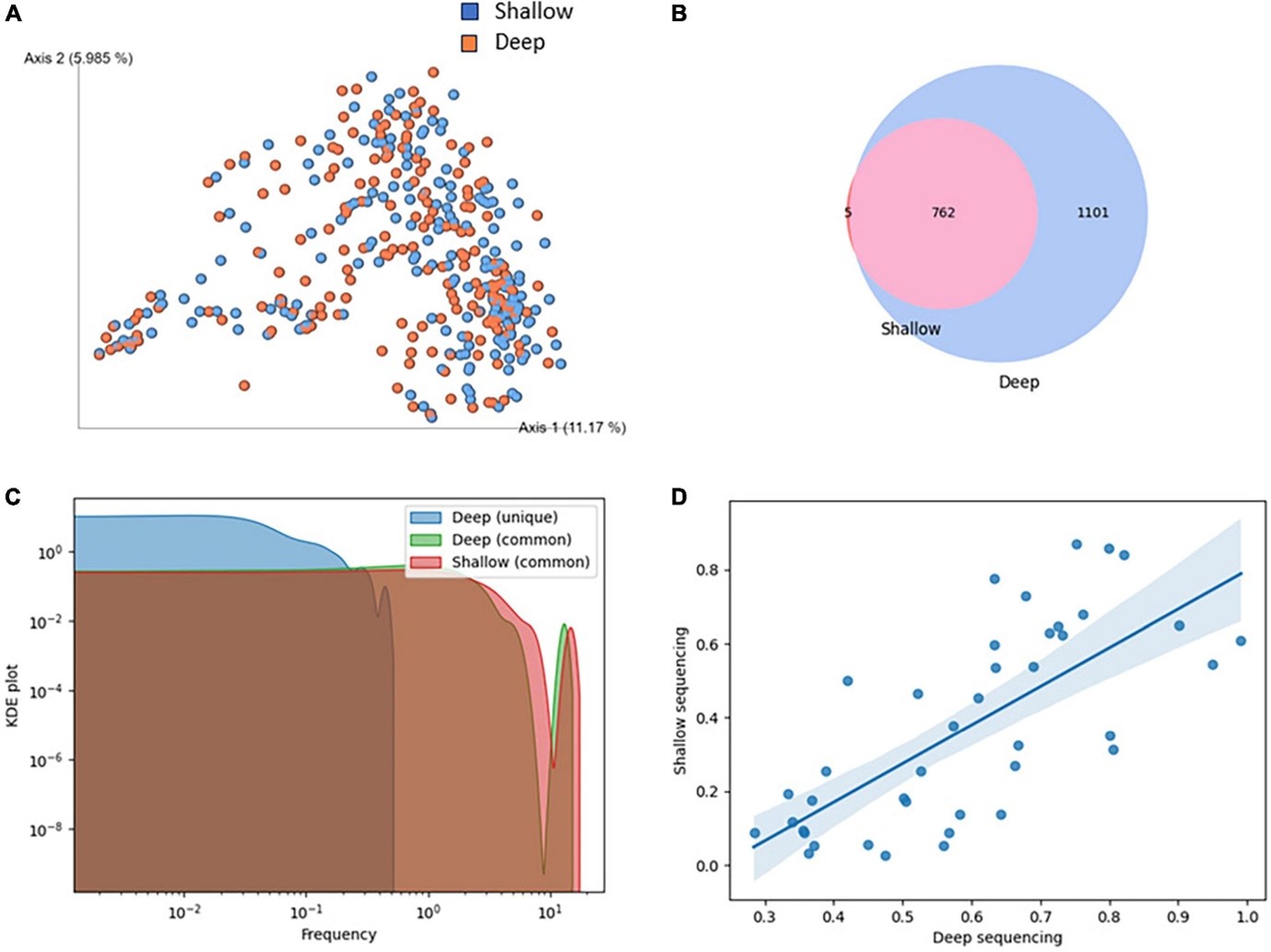
Figure 2. Comparison of shallow and deep shotgun sequencing methods. (A) Clustering of samples based on Bray-Curtis beta diversity. Blue: deep sequencing, red: shallow sequencing. (B) Intersection of species identified by shallow and deep sequencing. (C) KDE plot of abundance of species identified exclusively or jointly in shallow and deep sequencing.
The study was conducted by a team of MCB UJ researchers led by Dr. Tomasz Kościółek with support from Dr. Paweł Łabaj, in cooperation with Sanprobi sp. z o. o., Pomeranian Medical University in Szczecin and the National Medical Institute of the Ministry of Interior and Administration in Warsaw.
The uniqueness of the data and its abundance gives hope for further discoveries, especially with expanded analysis of the deep sequencing data. This analysis is currently being carried out by Dr. Tomasz Kościółek's Structural and Functional Genomics group (now: Sano - Center for Personalized Computational Medicine) with the participation of MCB UJ researchers and other consortium members.

