Proteomics and Mass Spectrometry Core Facility provides proteomic services to the researchers from the Jagiellonian University and to external users from academia and industry.
Urszula Jankowska, PhD
 Information about Core Facility Leader
Information about Core Facility Leader
Personal informations:
https://orcid.org/0000-0002-7165-8964
https://www.researchgate.net/profile/Urszula-Jankowska
https://www.linkedin.com/in/urszula-jankowska-69548884/
Dr Urszula Jankowska
Urszula Jankowska, PhD
Leader of Proteomics and Mass Spectrometry Core Facility
E-mail: urszula.jankowska@uj.edu.pl
Dr Bożena Skupień-Rabian
 Bożena Skupień-Rabian, PhD
Bożena Skupień-Rabian, PhD
Technician
E-mail: bozena.skupien-rabian@uj.edu.pl
Weronika Waluś
Weronika Waluś
Technician
E-mail: weronika1.walus@uj.edu.pl
Q Exactive (Thermo Scientific)
 Q Exactive (Thermo Scientific)
Q Exactive (Thermo Scientific)
It is a hybrid mass spectrometer, which combines high-performance quadrupole precursor selection with high-resolution, accurate-mass Orbitrap™ detection. Principally used for protein identification and quantification in complex samples (e.g. cell lysates), as well as identification of post-translational modifications.
micrOTOF-Q II (Bruker Daltonics)

micrOTOF-Q II (Bruker Daltonics)
It is used to measure molecular weight of intact proteins, peptides or nucleic acid fragments.
Proteomics and Mass Spectrometry Core Facility provides proteomic services to the researchers from the Jagiellonian University and to external users from academia and industry. We apply state-of-the-art methods for analyzing peptides and proteins, including their post-translational modifications (PTMs). We perform complete proteomic workflow that includes sample preparation, mass spectrometry measurement and data analysis. We also offer support in the conceptual design of experiments to apply the best strategy for each research question.
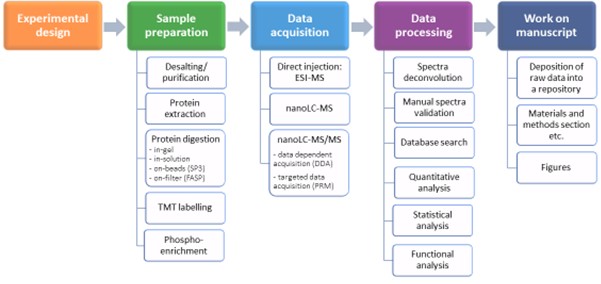
Figure 1. Scheme of the proteomic workflow and methodology available at the Proteomics and Mass Spectrometry CF. Among the variety of options, we recommend an optimal procedure fora given scientific question.
Proteomics is a comprehensive, large-scale analysis of the entire protein complement of a cell line, tissue, or organism. Mass spectrometry (MS) as a robust, sensitive and precise technique, enables the study of proteomes providing information on protein abundances, post-translational modifications and protein-protein interaction networks. In shotgun proteomics, proteins are first digested with protease, and then the resulting peptides are separated by liquid chromatography and analyzed by MS. As a result, in a large-scale proteomic experiment, thousands of different proteins can be identified and quantified, which can be utilized to i.a. characterize two biological states, elucidate mechanisms of disease, detect diagnostic markers or drug targets.
MS is also a powerful tool to analyze single protein providing mass determination of intact molecule, confirmation of protein identification, characterization of PTMs, detection of potential truncations and mutations.
Services offered by the Proteomics and Mass Spectrometry Core Facility include:
• Intact protein/peptide mass determination by ESI-MS
Service Guidelines (subpage: Guidelines_1_Mass_determination.doc)
Submission form (file: Form_1_Mass_determination.pdf)
• Protein identification from gel bands; characterization of post-translational modifications (PTMs) of single proteins
Service Guidelines (subpage: Guidelines_ProteinID_gel.doc)
Submission form (file: Form_ ProteinID_gel.pdf)
• Global proteome profiling – identification and/or relative quantification of proteins in complex mixtures
Service Guidelines (subpage: Guidelines_Global_proteomics.doc)
Submission form (file: Form_Global_proteomics.pdf)
• Protein-protein interaction (PPIs) analysis
Service Guidelines (subpage: Guidelines_Interactome.doc)
Submission form (file: Form_Interactome.pdf)
• Targeted proteomic analysis – data acquisition for predefined sets of peptides using parallel reaction monitoring (PRM) method
Please contact us directly.
Terms of use
All users are required to acknowledge the Core Facility if any data obtained in the facility has been used in posters, papers, and all other publications.
Please, use the wording as follows:
We acknowledge Proteomics and Mass Spectrometry Core Facility of the Malopolska Centre of Biotechnology, Jagiellonian University for mass spectrometry analysis.
Co-authorship
Apart from routine services, we also apply individual solutions to meet various research needs. Collaborative projects that require nonroutine sample preparation or extensive data analysis justifies a coauthorship on a manuscript that contains data generated in our facility.
Co-authorships :
2022:
Surman M, Kędracka-Krok S, Wilczak M, Rybczyński P, Jankowska U, Przybyło M. Comparative Proteomic Profiling of Ectosomes Derived from Thyroid Carcinoma and Normal Thyroid Cells Uncovers Multiple Proteins with Functional Implications in Cancer. Cells. 2022 Mar 31;11(7):1184. doi: 10.3390/cells11071184.
Ibáñez A, Skupien-Rabian B, Jankowska U, Kędracka-Krok S, Zając B, Pabijan M. Functional Protein Composition in Femoral Glands of Sand Lizards (Lacerta agilis). Molecules. 2022 Apr 6;27(7):2371. doi: 10.3390/molecules27072371.
Kubiak-Szymendera M, Skupien-Rabian B, Jankowska U, Celińska E. Hyperosmolarity adversely impacts recombinant protein synthesis by Yarrowia lipolytica-molecular background revealed by quantitative proteomics. Appl Microbiol Biotechnol. 2022 Jan;106(1):349-367. doi: 10.1007/s00253-021-11731-y.
2021:
Surman M, Kędracka-Krok S, Jankowska U, Drożdż A, Stępień E, Przybyło M. Proteomic Profiling of Ectosomes Derived from Paired Urothelial Bladder Cancer and Normal Cells Reveals the Presence of Biologically-Relevant Molecules. Int J Mol Sci. 2021 Jun 24;22(13):6816. doi: 10.3390/ijms22136816.
Pinski A, Betekhtin A, Skupien-Rabian B, Jankowska U, Jamet E, Hasterok R. Changes in the Cell Wall Proteome of Leaves in Response to High Temperature Stress in Brachypodium distachyon. Int J Mol Sci. 2021 Jun 23;22(13):6750. doi: 10.3390/ijms22136750.
Zacchini F, Heber MF, Arena R, Radczuk N, Jankowska U, Ptak GE. Perturbations of the hepatic proteome behind the onset of metabolic disorders in mouse offspring developed following embryo manipulation. Theriogenology. 2021 Sep 1;171:119-129. doi: 10.1016/j.theriogenology.2021.05.022.
Smejda M, Kądziołka D, Radczuk N, Krutyhołowa R, Chramiec-Głąbik A, Kędracka- Krok S, Jankowska U, Biela A, Glatt S. Same but different - Molecular comparison of human KTI12 and PSTK. Biochim Biophys Acta Mol Cell Res. 2021 Apr;1868(4):118945. doi: 10.1016/j.bbamcr.2020.118945.
2020:
Pabis M, Termathe M, Ravichandran KE, Kienast SD, Krutyhołowa R, Sokołowski M, Jankowska U, Grudnik P, Leidel SA, Glatt S. Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation. EMBO J. 2020 Oct 1;39(19):e105087. doi: 10.15252/embj.2020105087.
Publications with Core Facility support:
2022:Matsuda A, Plewka J, Chykunova Y, Jones AN, Pachota M, Rawski M, Mourão A, Karim A, Kresik L, Lis K, Minia I, Hartman K, Sonani R, Schlauderer F, Dubin G, Sattler M, Suder P, Popowicz P, Pyrć K, Czarna A. Despite the odds: formation of the SARS-CoV-2 methylation complex. bioRxiv. 2022
2021:
Kumar M , Markiewicz-Mizera J, Janna Olmos JD, Wilk P, Grudnik P, Biela AP, Jemioła-Rzemińska M, Górecki A, Chakraborti S, Heddle JG. A single residue can modulate nanocage assembly in salt dependent ferritin. Nanoscale. 2021 Jul 15;13(27):11932-11942. doi: 10.1039/d1nr01632f.